Adapted from Science 2001 291: 1221
I. Chemical Proteomics
Chemical proteomics involves the principles of chemistry and proteomics to unravel the intricate mechanisms within biological systems. Typically, chemical proteomics is a powerful approach that employs small molecules as probes to scrutinize the vast and dynamic landscape of proteins, elucidating their functions, interactions, and regulatory mechanisms. Unlike traditional proteomics, which primarily relies on mass spectrometry and other analytical techniques, chemical proteomics introduces a chemical dimension by using chemical probes to selectively tag and manipulate proteins within their native cellular environment. This methodology allows researchers to not only identify and quantify proteins but also to interrogate their specific roles in signaling pathways, cellular processes, and disease states. The integration of chemical tools into proteomic studies enables a more comprehensive understanding of the functional proteome, paving the way for the discovery of novel drug targets, elucidation of biological mechanisms, and the development of personalized therapeutic interventions.
Study I: Tracking Virus EntryThe outbreak of Zika virus (ZIKV) in 2016 created worldwide health emergency which demand urgent research efforts on understanding the virus biology and developing therapeutic strategies. Here, we present a time-resolved chemical proteomic strategy to track the early-stage entry of ZIKV into host cells. ZIKV was labeled on its surface with a chemical probe, which carries a photocrosslinker to covalently link virus-interacting proteins in living cells on UV exposure at different time points, and a biotin tag for subsequent enrichment and mass spectrometric identification of the receptor or other host proteins critical for virus internalization. We identified Neural Cell Adhesion Molecule (NCAM1) as a potential ZIKV receptor and further validated it through overexpression, knockout, and inhibition of NCAM1 in Vero cells and human glioblastoma cells U-251 MG. Collectively, the strategy can serve as a universal tool to map virus entry pathways and uncover key interacting proteins.
II. Extracellular vesicle and Biomarker discovery
Our biomarker discovery projects apply proteomic strategies to detect extracellular vesicles (EV) proteomes derived from biofluids (plasma, urine, saliva) in an effort to find candidate markers of diseases like cancers, Pakinson’s and Alzheimer’s diseases.
Blood tests, which are common diagnostic procedures in clinical analyses, find blood biomarkers to categorize patients and make treatment decisions. However, existing biomarkers often lack specificity and are far from comprehensive and may hinder diagnoses. Mass spectrometry (MS)-based proteomics has become a powerful tool in the biomarker discovery area, as it allow us to characterize biofluid proteins in great depth and specificity.
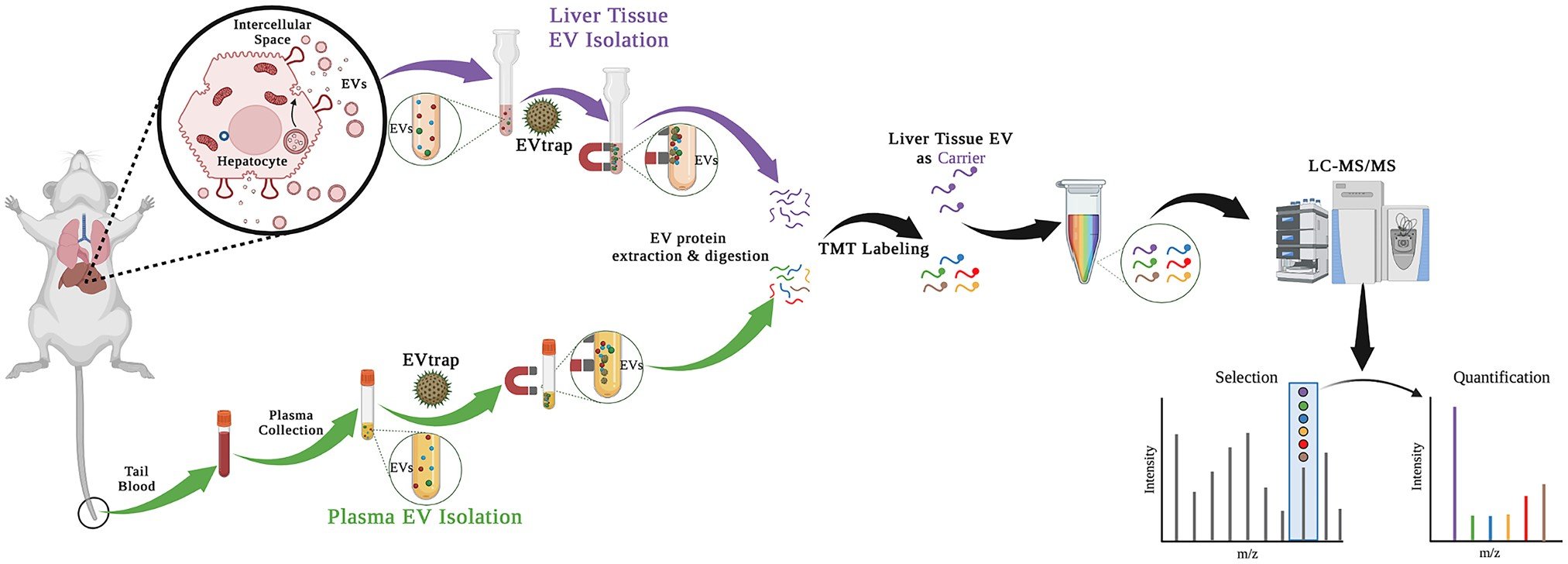
Study I: Drug Metabolism Monitoring through EVsThe ability to monitor the response of metabolic enzymes to drug exposure in individuals is highly appealing and critical to personalized medicine. Here, we report a quantitative proteomics strategy to monitor drug metabolic pathways by profiling metabolic enzymes in circulating extracellular vesicles (EVs) upon drug exposure. Mass spectrometry (MS)-based measurement revealed that changes in metabolic enzyme abundance in EVs paralleled those in hepatic cells isolated from liver tissue. Coupling with multiplexed isotopic labeling, we temporally quantified 34 proteins involved in drug absorption, distribution, metabolism, and excretion (ADME) pathways. Out of 44 known ADME proteins in plasma EVs, previously annotated mouse cytochrome P450 3A11 (Cyp3a11), homolog to human CYP3A4, and uridine 5’-diphospho (UDP) glucuronosyltransferase 2A3 (Ugt2a3), increased upon daily rifampicin dosage. Altogether, this study demonstrates that measuring drug enzymes in circulating EVs as an effective surrogate is highly feasible and may transform today’s drug discovery and development for personalized medicine.
Study II: Diagnostic Biomarkers for Parkinson’s DiseaseMutations in the leucine-rich repeat kinase 2 (LRRK2) gene have been recognized as genetic risk factors for Parkinson’s disease (PD). However, compared to cancer, fewer genetic mutations contribute to the cause of PD, propelling the search for protein biomarkers for early detection of the disease. Utilizing 138 urine samples from four groups, healthy individuals (control), healthy individuals with G2019S mutation in the LRRK2 gene (non-manifesting carrier/NMC), PD individuals without G2019S mutation (idiopathic PD/iPD), and PD individuals with G2019S mutation (LRRK2 PD), we applied a phosphoproteomics strategy to determine potential diagnostic biomarkers for PD from urinary extracellular vesicles (EVs). The findings in this study demonstrate a general strategy of utilizing biofluid EV proteome/phosphoproteome as an outstanding and non-invasive source for a wide range of disease exploration.
Study III: One-Pot Proteomic Analysis of EVsThe current workflow of mass spectrometry-based EV proteome analysis is not fully compatible in a clinical setting due to inefficient EV isolation methods and a tedious sample preparation process. To streamline and improve the efficiency of EV proteome analysis, we introduced a one-pot analytical pipeline integrating a robust EV isolation approach, EV total recovery and purification (EVtrap), with in situ protein sample preparation, to detect urinary EV proteome. By incorporating solvent-driven protein capture and fast on-bead digestion, the one-pot pipeline enabled the whole EV proteome analysis to be completed within one day. Our novel one-pot analytical pipeline demonstrated its potential for routine and robust EV proteomics in biomedical applications.
III. protein post translational modifications
Besides the proteins, protein post-translational modifications (PTMs) are also interesting targets because of their involvement in many disease-related signaling pathways. Our current pipeline focuses on an unbiased, comprehensive discovery phase to identify biomarker candidates resulting from disease associated differential expression and aberrant PTMd (phosphorylation, acetylation, glycosylation) proteins. We also developed in vitro assays to identify and validate top priority possible targets. Moreover, our group has been working on the development of various technologies for PTM-related detection.
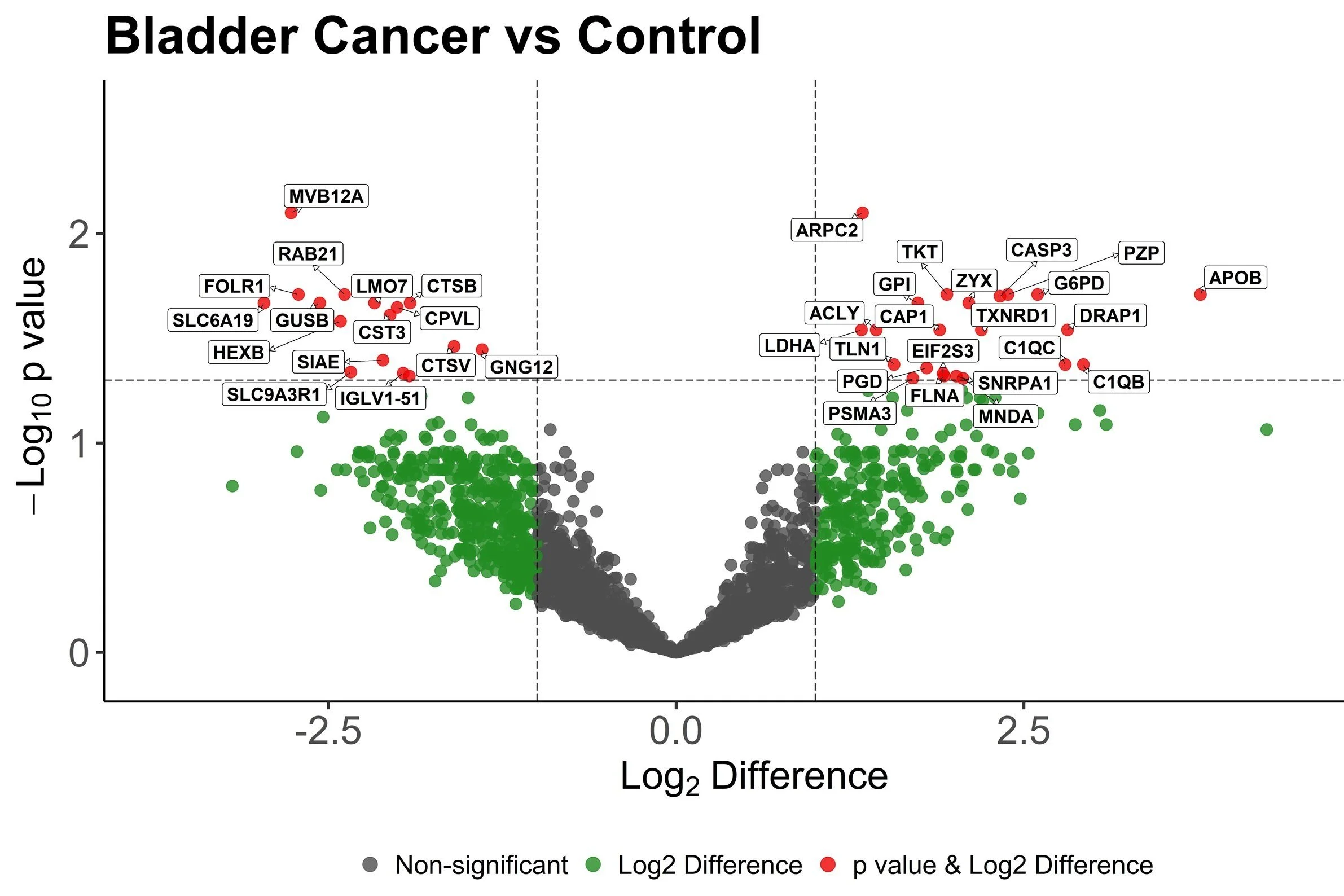
Study I: Distinguishing Tumor Grades with PhosphoproteomicsIn this study, urinary extracellular vesicular proteins are compared between healthy controls, patients with chronic kidney disease, and patients with either low grade or high-grade renal cell carcinoma (RCC). Knowing the grade of renal cell carcinoma is critical knowledge for surgeons recommending operations, but existing determination methods rely on painful and invasive tumor biopsies. The developed liquid biopsy for distinguishing low and high grade RCC utilizes urinary extracellular vesicular enrichment via EVtrap and phosphoprotein enrichment with PolyMac beads. After mass spectral analysis, upregulated phosphorylation was shown to be site specific between patients suffering from low-grade versus high-grade RCC, indicating that this technique is viable for distinguishing RCC grades even when compared to other kidney diseases or healthy patients.
Study II: Kinase Assay Linked PhosphoproteomicsProtein kinases are major regulatory components in almost all cellular processes in eukaryotic cells. By adding phosphate groups, protein kinases regulate the activity, localization, protein–protein interactions, and other features of their target proteins. It is known that protein kinases are central components in plant responses to environmental stresses such as drought, high salinity, cold, and pathogen attack. However, only a few targets of these protein kinases have been identified. Moreover, how these protein kinases regulate downstream biological processes and mediate stress responses is still largely unknown. In this study, we introduce a strategy based on isotope-labeled in vitro phosphorylation reactions using in vivo phosphorylated peptides as substrate pools and apply this strategy to identify putative substrates of nine protein kinases that function in plant abiotic and biotic stress responses.